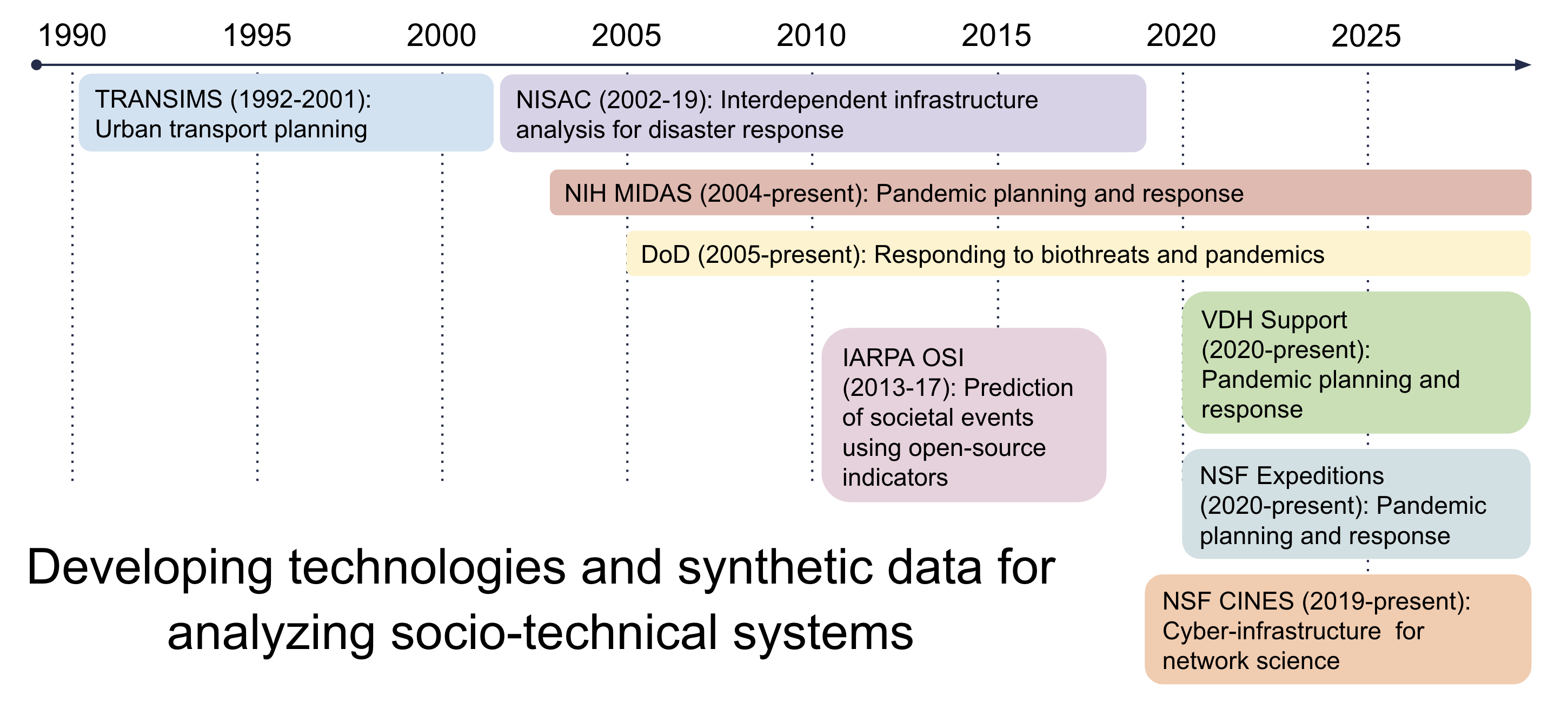
NSSAC Research
Supporting Collaborative Research

Since our team wrote this influential Nature paper in 2004, we have supported US government agencies in their work to address public health emergencies, including planning for the H5N1 outbreak, the 2009 H1N1 pandemic, the MERS outbreak of 2012, the 2014 Ebola outbreak, the 2016 Zika outbreak, the most recent Ebola outbreak in Congo, and since January 2020 we’ve been supporting several local, state, and federal agencies in the fight against COVID-19.
These are some of the publications that help outline our work:
Eubank S, Guclu H, Vullikanti A, Marathe M, Srinivasan A, Toroczkai Z, Wang N. Modelling disease outbreaks in realistic urban social networks. Nature. 2004 May;429(6988):180-4. Link
Adiga A, Dubhashi D, Lewi B, Marathe M, Venkatramanan S, Vullikanti A. Mathematical models for COVID-19 pandemic: a comparative analysis. Journal of the Indian Institute of Science, 2020 Oct;100(4):793-807. Link
Adiga A, Chen J, Marathe M, Mortveit H, Venkatramanan S, Vullikanti A. Data-driven modeling for different stages of pandemic response. Journal of the Indian Institute of Science, 2020 Oct;100(4):901-15. Link
Chen J, Levin S, Eubank S, Mortveit H, Venkatramanan S, Vullikanti A, Marathe M. Networked epidemiology for COVID-19. SIAM News. 2020 Jun;53(5). Link
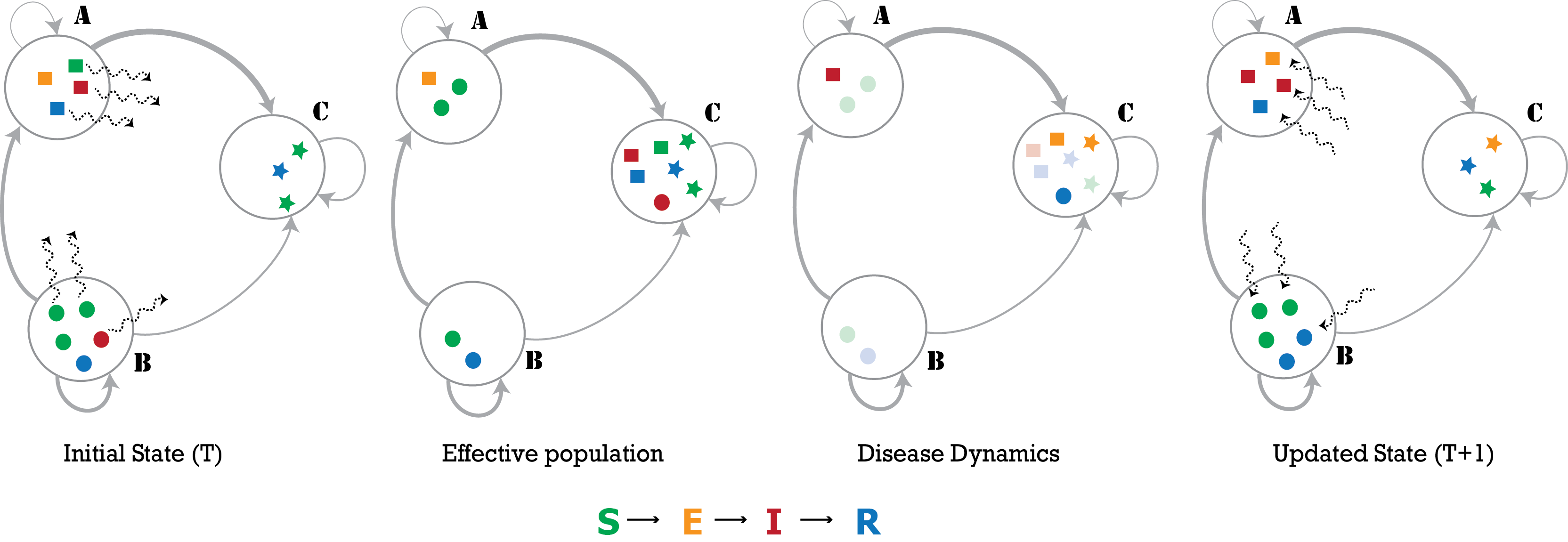
We have developed a national-scale metapopulation model for the spread of influenza by integrating both local and long-distance mobility data. For the example below, we combined data on commuter mobility from the American Community Survey (ACS) with domestic airline passenger data from the Bureau of Transportation Statistics (BTS) to capture human mobility across the country. Next, we adopted a metapopulation approach to simulate epidemic spread at the spatial resolution of counties (patches) wherein the temporal travel matrix is used to represent the flow of people between these patches: PatchSim.
Venkatramanan S, Chen J, Fadikar A, Gupta S, Higdon D, Lewis B, Marathe M, Mortveit H, Vullikanti A. Optimizing spatial allocation of seasonal influenza vaccine under temporal constraints. PLoS Computational Biology. 2019 Sep 16;15(9):e1007111. Link
PatchSim has subsequently been used in this Nature Communications article for influenza forecasting.
Venkatramanan S, Sadilek A, Fadikar A, Barrett C, Biggerstaff M, Chen J, Dotiwalla X, Eastham P, Gipson B, Higdon D, Kucuktunc O. Forecasting influenza activity using machine-learned mobility map. Nature Communications. 2021 Feb 9;12(1):1-2. Link
Click here to start using PatchSim.
 PatchSim can be used in modeling a variety of situations. Click the links below to learn about:
PatchSim can be used in modeling a variety of situations. Click the links below to learn about:
We developed this PatchSim dataset for our project COVID-19 Response Support: Building Synthetic Multi-scale Networks, funded through the NSF RAPID program. This material is based upon work supported by the National Science Foundation under Grant No. OAC-2027541.
Rajuladevi R, Marathe M, Porebski P, Venkataramanan S. Impact of Seeding and Spatial Heterogeneity on Metapopulation Disease Dynamics. Proceedings of epiDAMIK 5.0: The 5th International Workshop on Epidemiology meets Data Mining and Knowledge Discovery at KDD 2022. Link
This work was supported by NSF RAPID 2027541: COVID-19 Response Support: Building Synthetic Multi-scale Networks, NSF Expeditions 1918656: Collaborative Research: Global Pervasive Computational Epidemiology, and DTRA subcontract/ARA S-D00189-15-TO01-UVA.
Other PatchFlow datasets use GPWv4 and GADM v3.6
EpiHiper is a parallel agent-based socio-epidemic simulator. In a distributed network-based stochastic disease transmission simulation, it allows you to assess the impact on transmission under different conditions and assess the impact of contact tracing.
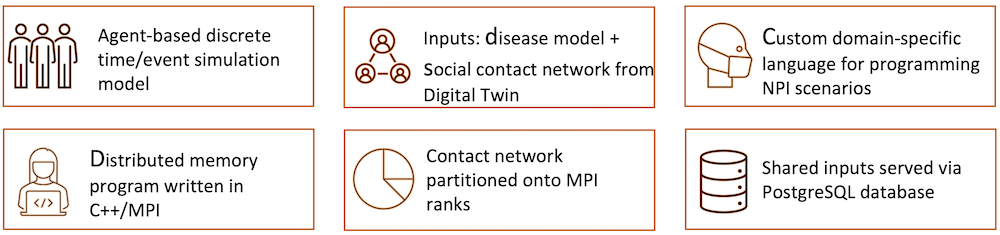
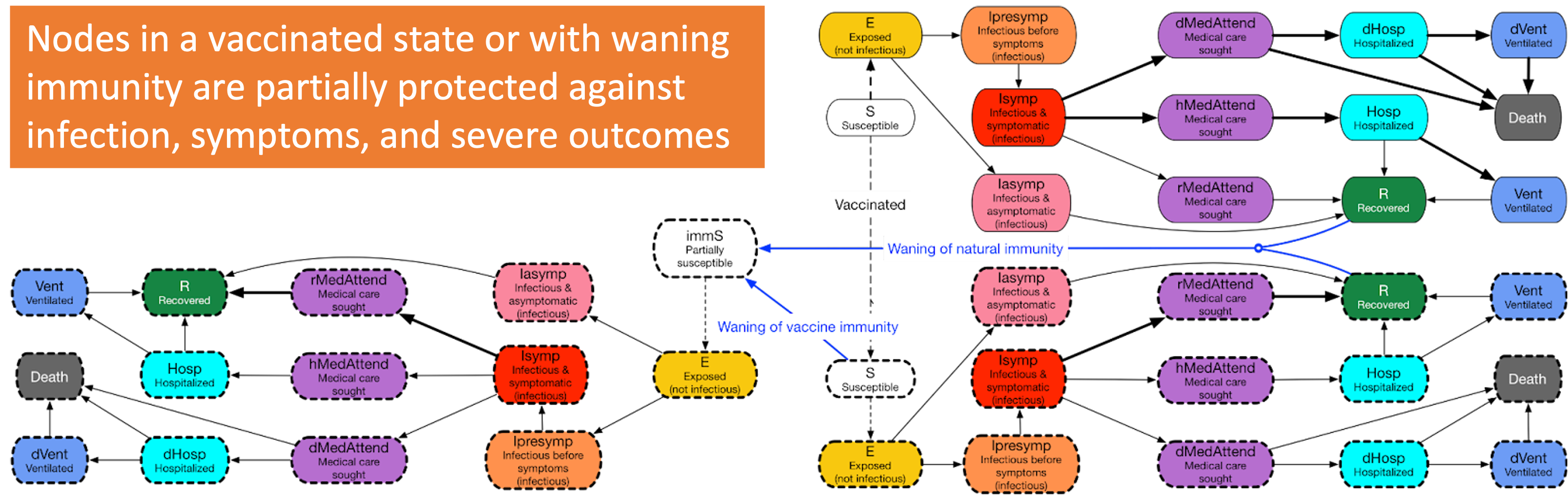
A great example is our use of EpiHiper in running a disease model with vaccination and waning immunity.
You can learn more about the way we’ve used EpiHiper by clicking the links below:
Our paper Data-Driven Scalable Pipeline using National Agent-Based Models for Real-time Pandemic Response and Decision Support on the innovative creation of a high performance computing pipeline was named a finalist for the 2021 ACM Gordon Bell Special Prize for High Performance Computing-Based COVID-19 Research. Read more about this special prize in Nature Computational Science.
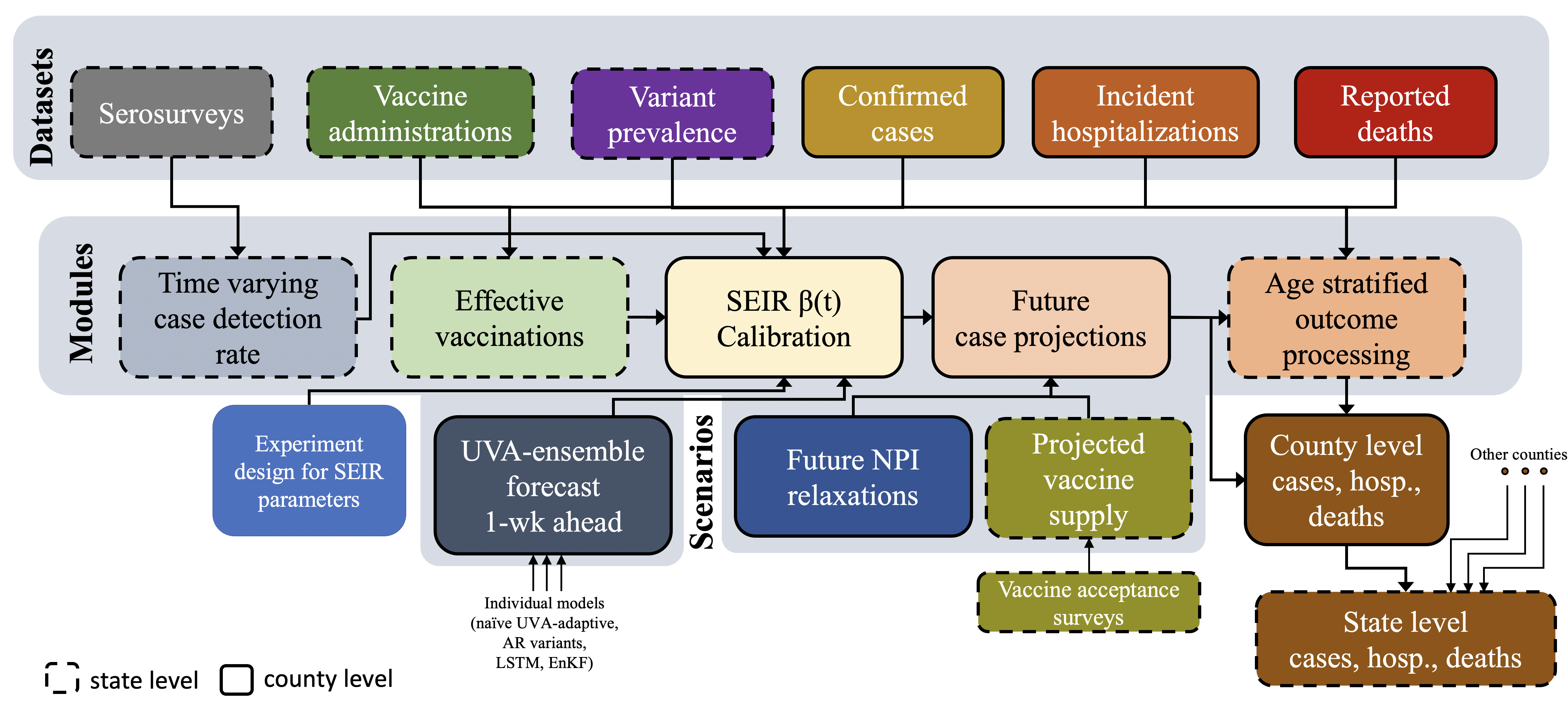
We have developed a multi-scale, multi-target forecasting framework for providing short-term, real-time forecasts of epidemic cases and hospitalizations for illnesses like influenza and COVID-19. The model is flexible and has been deployed at multiple resolutions for the forecasting of state-level COVID-19 hospitalizations and COVID-19 incident cases at the US national, state, and county level. Additionally, we provide forecasts for European countries and Indian states.
We employ multiple classes of methods: (i) statistical – Autoregressive and Kalman filters; (ii) a Deep learning model – (Long-short term memory); and (iii) compartmental models (a variant of PatchSim). The forecasts from the individual models are ensembled using Bayesian Model Averaging. The model provides both point and probabilistic forecasts. The influenza model also allows for human-judgement-ensembling through voting.

The COVID-19 model forecasts have been submitted on a weekly basis to the CDC Forecast Hub and the European CDC Forecast Hub for over two years.
The weekly influenza hospitalization forecasts for the 2021-2022 season were submitted to the CDC FluSight for inclusion in their ensemble forecasts.
You can find our dashboard for the visualization and inspection of multi-target, multi-resolution forecasts here.
More details about the model are provided in: Adiga A, Wang L, Hurt B, Peddireddy A, Porebski P, Venkatramanan S, Lewis B, Marathe M. All models are useful: Bayesian ensembling for robust high resolution COVID-19 forecasting. Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery & Data Mining, 2021 Aug 14;2505-2513. Link
Details about the COVID-19 Forecast Hub forecasts and contributing teams is provided in:
Cramer EY, Huang Y, Wang Y, Ray EL, Cornell M, Bracher J, Brennen A, Rivadeneira AJ, Gerding A, House K, Jayawardena D, et al. The United States COVID-19 Forecast Hub dataset. Scientific Data. 2022 Aug 1;9(1):1-5. Link
The performance of the ensemble and the forecasts from contributing teams is discussed in:
Sherratt K, Gruson H, Johnson H, Niehus R, Prasse B, Sandman F, Deuschel J, Wolffram D, Abbott S, Ullrich A, Gibson G, et al. Predictive performance of multi-model ensemble forecasts of COVID-19 across European nations. medRxiv. 2022 Jun 16. DOI 10.1101/2022.06.16.22276024v1. (Submitted to PLOS Global Public Health) Link